With every article and podcast episode, we provide comprehensive study materials: References, Executive Summary, Briefing Document, Quiz, Essay Questions, Glossary, Timeline, Cast, FAQ, Table of Contents, Index, Polls, 3k Image, and Fact Check.
The world barely had time to process the mRNA vaccines that helped us through a pandemic when scientists just quietly dropped another bombshell. This one didn't make headlines, but it should have.
In November 2024, researchers at Nagoya University published findings that could fundamentally transform how we treat diseases ranging from rare genetic disorders to common killers. While most of us were doom-scrolling through election coverage, they were solving a problem that has challenged medical science for decades.
The Problem with Traditional mRNA
Let's get something straight: traditional linear mRNA treatments are remarkable, but they're fundamentally flawed. They degrade quickly inside the body, like messages written on dissolving paper. This is why current mRNA therapies often require multiple doses and careful handling. The body's cellular machinery reads these molecules once, produces the needed protein, and then the mRNA rapidly breaks down.
Linear mRNA is the medical equivalent of planned obsolescence. It does its job, but inefficiently, requiring patients to return for multiple treatments while pharmaceutical companies enjoy recurring revenue. This isn't a conspiracy; it's just the biological reality of how these molecules function.
Enter Circular mRNA: The Sustainable Alternative
Circular mRNA is different. By forming a continuous loop without exposed ends, it resists degradation by the cell's cleanup enzymes. This provides longevity that linear mRNA can't match – potentially lasting weeks instead of hours or days.
But there's been a critical barrier: circular mRNA lacks the specialized "cap" structure that linear mRNA uses to engage with cellular machinery. Without this cap, protein production becomes inefficient – like having an everlasting light bulb that barely glows.
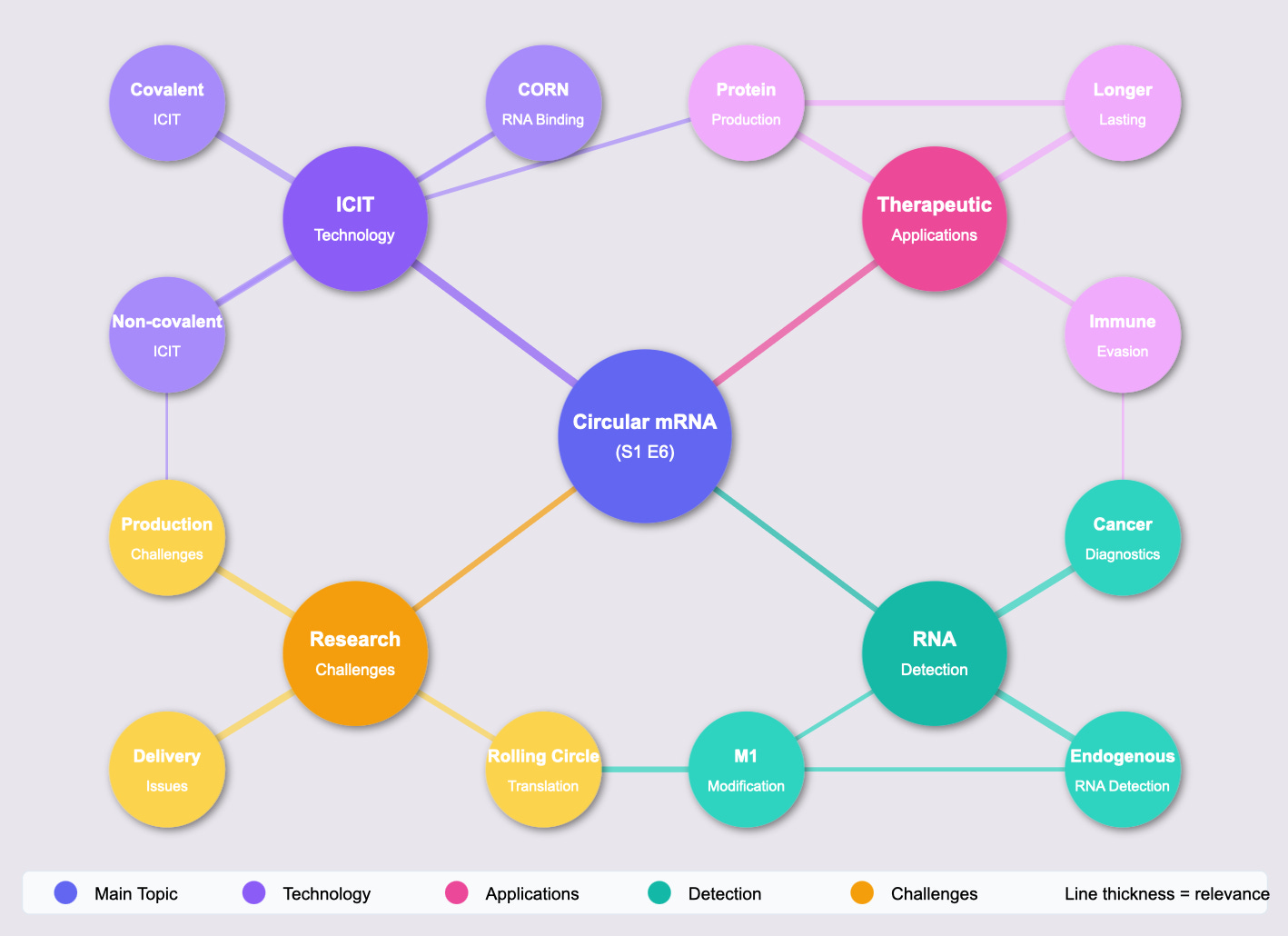
The brilliance of the Nagoya researchers lies in how they solved this problem. Through a technique they call "Internal Cap-Initiated Translation" (ICIT), they devised two methods to equip circular mRNA with the necessary cap structure:
Covalent ICIT: Directly attaching a cap to a branch extending from the RNA loop
Non-covalent ICIT: Using separate "CORN" RNA molecules that carry the cap and bind to the circular mRNA
The results are staggering. The covalent approach yielded a 100-fold increase in protein production compared to uncapped circular mRNA. The non-covalent method achieved a 50-fold increase. These aren't incremental improvements – they're transformative leaps that could make treatments dramatically more effective while requiring fewer doses.
What This Actually Means For People
When scientists talk about "protein production efficiency," eyes glaze over. But here's what this really means for human beings:
For patients with chronic conditions like hemophilia or certain enzyme deficiencies, this could mean monthly treatments instead of weekly ones, or quarterly instead of monthly. It means fewer disruptions to life, less time in medical facilities, and reduced treatment burden.
For people fighting cancer, it could lead to more potent immunotherapies that train the body to recognize and destroy tumor cells more effectively and for longer periods from a single treatment.
For the healthcare system, it represents potentially massive cost savings. Fewer doses mean lower manufacturing costs, reduced cold-chain logistics requirements, and fewer clinical visits.
But perhaps most exciting is the discovery of "rolling circle translation" – where a single circular mRNA molecule can serve as a template to produce multiple protein copies in succession. This is like having a printing press that never needs new plates, continuously producing exactly what the body needs.
The Diagnostic Revolution Nobody Saw Coming
Beyond therapeutic applications, the researchers demonstrated something equally revolutionary: using their "CORN" technology to detect specific RNAs within cells. They showed they could identify a long non-coding RNA called HULC that's elevated in certain cancers, as well as detect natural cellular RNAs like beta-actin.
Think about what this means: we could potentially develop diagnostic tools that detect the earliest signs of cancer or monitor treatment effectiveness in real-time. The same technology treating disease could simultaneously monitor the disease's presence.
This convergence of treatment and diagnostics – "theranostics" as some researchers call it – represents a new paradigm in medicine that moves beyond treating symptoms to addressing the most fundamental molecular causes of disease with unprecedented precision.
Why Aren't We Hearing More About This?
The silence surrounding this breakthrough is deafening. While celebrities and politicians dominate news cycles, potentially life-changing scientific developments receive minimal attention. The reasons are manifold:
These advances are complex and difficult to communicate to general audiences
They represent early-stage research that hasn't yet been commercialized
The full impact won't be felt for years as the technology moves through development and regulatory approval
But make no mistake – this is the kind of fundamental scientific advancement that transforms medicine. Similar to how the development of monoclonal antibodies in the 1970s led to revolutionary treatments decades later, today's circular mRNA breakthroughs could reshape healthcare delivery in the 2030s and beyond.
The Challenges Ahead
Scientific breakthroughs always face obstacles on the path to real-world impact. For circular mRNA technologies, several challenges remain:
Manufacturing complexity: Creating branched, capped RNA molecules at scale requires sophisticated techniques that may be difficult to industrialize initially.
Delivery mechanisms: Getting these molecules into the right cells remains a significant hurdle, though lipid nanoparticle technology has advanced substantially in recent years.
Regulatory pathways: Novel therapeutic modalities face rigorous scrutiny from regulatory bodies, requiring extensive testing to demonstrate safety and efficacy.
However, these are engineering and regulatory challenges, not fundamental scientific barriers. The core breakthrough – enabling efficient protein production from circular mRNA – has been achieved.
The Bigger Picture
This research represents more than just another incremental advance in biotechnology. It exemplifies how scientific progress often occurs through creative problem-solving at the molecular level, with implications that ripple outward to affect millions of lives.
When we look back at the evolution of medicine, we'll likely see this moment as pivotal – when RNA-based therapeutics moved from being remarkable but limited tools to becoming sustainable platforms for treating a vast array of conditions.
The future of medicine isn't just about developing new drugs; it's about creating more efficient, precise, and sustainable ways to deliver therapeutic effects. Circular mRNA technology embodies this shift toward treatments that work with the body's own machinery to produce lasting benefits from minimal intervention.
As we face growing healthcare costs and increasing chronic disease burden, innovations that reduce treatment frequency while maintaining or improving efficacy aren't just scientifically impressive – they're economically and socially essential.
What Happens Next?
The path from laboratory discovery to approved treatment typically spans 10-15 years, but RNA technologies have moved with unprecedented speed in recent years. The COVID-19 pandemic demonstrated how quickly mRNA platforms can be deployed when necessary.
In the next few years, expect to see:
Additional research refining and optimizing these technologies
Partnerships between academic institutions and biotechnology companies to advance development
Initial clinical trials for specific applications where circular mRNA offers clear advantages
The most promising early applications will likely target rare diseases where conventional treatments are inadequate, expensive, or burdensome. Success in these areas will pave the way for broader applications.
The Bottom Line
While much of the world remains distracted by the immediate and the trivial, scientists continue making remarkable progress on challenges that could reshape human health. The circular mRNA breakthroughs from Nagoya University won't make headlines like celebrity gossip or political scandals, but their long-term impact on human wellbeing will likely be immeasurably greater.
The future of medicine is being written now, not in hospital corridors or pharmaceutical boardrooms, but in laboratories where researchers are reimagining the most fundamental tools we use to combat disease. And that future looks increasingly circular.
Link References
The Circular RNA Revolution: Why This Matters More Than You Think (S3 E31) HelioxPodcast: Where Evidence Meets Empathy
Reference:
Internal cap-initiated translation for efficient protein production from circular mRNA
Episode:
The Circular RNA Revolution: Why This Matters More Than You Think (S3 E31)
Podcast:
Heliox: Where Evidence Meets Empathy
Substack:
Select your podcast provider
Heliox: Where Evidence Meets Empathy on Youtube
STUDY MATERIALS
1. Briefing Document
Source: Excerpts from "Internal_cap-initiated_translation_provides_effici.pdf" (Preprint posted November 5th, 2024)
Authors: Hiroshi Abe et al., Nagoya University and other institutions.
Key Themes:
This preprint introduces two novel molecular designs to significantly enhance protein production from circular messenger RNA (circRNA) by employing an internal cap-initiated translation (ICIT) mechanism. This approach aims to overcome the limitations of traditional IRES (internal ribosome entry site)-dependent translation in circRNAs, offering improved efficiency, smaller molecular size, and the potential for cell-type selective translation and RNA detection.
Most Important Ideas and Facts:
Challenges with Circular mRNA Translation: While circRNAs offer benefits like increased stability and reduced immunogenicity compared to linear mRNAs, they lack the 5' cap structure essential for efficient cap-dependent translation. Current methods often rely on IRES elements, which exhibit intrinsically lower translation efficiency.
"Circular mRNA, emerging as a groundbreaking RNA therapeutic strategy, faces challenges in enhancing its translation potential. Without an N7-methylguanosine (m7G) structure, circular RNAs require atypical translation initiation mechanisms." "However, the intrinsically low eciency of IRES-mediated translation initiation hampers its pharmacological applications."
Two Novel ICIT Strategies: The study presents two distinct methods to internally position an m7G cap on circRNAs to promote translation:
Covalent Attachment (cap-circ mRNA): This design involves a circRNA with an m7G cap covalently attached through a branched RNA structure incorporated into the 5' UTR.
"The rst design involved a circular mRNA with a covalently attached N7-methylguanosine (m7G) cap through a branching structure (cap-circ mRNA). This modication allows circular mRNA to recruit translation machinery and produce proteins more eciently than IRES-containing circular mRNAs."
Non-covalent Attachment (cORN-mediated): This strategy utilizes a short, m7G cap-containing oligonucleotide (cORN) that hybridizes to a complementary sequence within the circRNA, effectively tethering the cap to the circular molecule.
"The second design features the non-covalent attachment of an m7G cap to a circular mRNA through hybridization with an m7G cap-containing oligonucleotide, signicantly enhancing translation by more than 50-fold."
Enhanced Translation Efficiency of cap-circ mRNA: Covalent introduction of the m7G cap dramatically increased protein expression levels (two-to-three orders of magnitude) compared to uncapped circRNAs. This translation was confirmed to be dependent on canonical cap-dependent translation initiation factors (eIF4E, eIF4A, and PABP).
"Circular mRNAs possessing a capped branched strand (CircR3 and CircR9) exhibited ecient Nluc expression with a 103-fold increase for CircR3 compared to CircR2 and 189-fold increase for CircR9 compared to CircR8." "Therefore, the translation initiation process of cap-circ mRNA may utilize conventional factors for cap-dependent translation, including eIF4E, eIF4A, and PABP."
Improved Stability and In Vivo Performance of cap-circ mRNA: cap-circ mRNA demonstrated more durable protein expression in mice compared to linear mRNA, likely due to its inherent stability. Combining the cap-circ design with N1-methylpseudouridine (m1Ψ) modification further increased translation efficiency and reduced acute immunostimulatory effects in mice, showing comparable or superior performance to optimized IRES-containing circRNAs in vivo.
"We conrmed that the protein expression of cap-circ mRNA was more durable than that of linear mRNA in vivo mouse studies." "Combining N1-methylpseudouridine (m1Ψ) modication, cap-circ mRNA exhibits a lower acute immunostimulatory effect, maintaining high translation ability, in mice." "In both administration routes, cap-circ m1Ψ mRNA outperformed HRV-B3-IRES-containing circ mRNA."
Smaller Molecular Size: The cap-circ strategy requires only a short capped RNA fragment (a few dozen bases) compared to the much larger IRES sequences (several hundred bases), resulting in smaller overall circRNA size. This can be advantageous for delivery and potentially translation efficiency, especially for short peptides.
"Additionally, our strategy can increase the translation eciency of circular mRNA with a minimal increase in its length of circular mRNA, providing another practical advantage. Capped fragments are only a few dozen bases long, whereas IRES sequences typically consist of several hundred bases."
Non-covalent ICIT via cORNs: Hybridization of m7G-capped oligoribonucleotides (cORNs) to circRNAs significantly enhanced translation (more than 50-fold) in a hybridization-dependent manner, even in the absence of IRES elements. The position and length of the cORN, as well as chemical modifications (like a 3' poly(A) tail), influenced the degree of translation enhancement.
"Intriguingly, pre-annealing of the 20-nt cORNs to the circular mRNA increased translation from the circular mRNA, even without IRES or capped branch strands... Hybridization to the most distant site (Target site-1) provided the highest potential for translation enhancement among the three target sites." "A poly A tail (A30) conjugated to the 3′ end of cORN (cORN-14) increased translation by 1.5-fold..."
Applications of cORN-mediated Translation: This non-covalent strategy opens up possibilities for:
Rolling Circle Translation: cORNs can facilitate the initiation of rolling circle-type translation from circRNAs lacking stop codons, leading to the production of long, tandemly linked protein products.
"Harnessing cORN, we found that this system can also facilitate rolling circle-type translation of circular mRNA... In vitro translation of circular mRNA showed that hybridization with capped cORN (cORN-19) increased the production eciency of large proteins with tandem repeats of FLAG-EGF..."
Detection of Cellular RNAs (Translation Switches): By designing circRNAs with sequences complementary to capped cellular RNAs (including lncRNAs and mRNAs), the presence of the target RNA can trigger translation of a reporter protein from the circRNA by "borrowing" the cap. This was demonstrated with both exogenously introduced and endogenous RNAs like HULC lncRNA and hACTB mRNA.
"Here we applied this system to long non-coding RNAs (lncRNAs)... Here, we investigated whether cap-structured RNAs led to the enhancement of the circular mRNA translation... As expected, HULC RNA increased the translation of circular mRNA by 18-fold cap-dependently." "This result suggests that endogenous hACTB mRNA induces the translation of circular mRNAs."
Comparison of Covalent and Non-covalent ICIT: While covalent ICIT (cap-circ mRNA) generally exhibited higher translation efficiency, the non-covalent cORN approach offers simplicity in nucleic acid synthesis and versatility for applications like RNA detection.
"We note that internal cap introduction (i.e., covalent ICIT) was still a better option to drive protein from circular mRNAs than hybridizing cORN (i.e., non-covalent ICIT)... Nonetheless, considering the simplicity of nucleic acid synthesis in industry, cORN could be a useful strategy for therapeutic applications."
Potential Competing Interests:
Hiroshi Abe (HA) and Satomi Sugiyama (SU) are cofounders of Crafton Biotechnology, which focuses on mRNA therapeutics.
An international patent application covers part of this work.
Shintaro Iwasaki (SI) is a member of the Scientific Reports editorial board.
Conclusion:
This research presents significant advancements in the field of circular RNA technology by introducing two effective internal cap-initiated translation strategies. These methods address key limitations of circRNA-based therapeutics and diagnostics, offering enhanced translation efficiency, smaller molecular size, and novel functionalities for RNA detection and potentially cell-type specific protein expression. The findings have promising implications for the development of next-generation mRNA vaccines and therapeutics.
2. Quiz & Answer Key
Study Guide: Internal Cap-Initiated Translation of Circular mRNA
Quiz
What is the primary challenge in utilizing circular mRNA for therapeutic applications, and how do traditional methods like IRES address this?
Describe the two main innovative molecular designs introduced in this study to enhance circular mRNA translation.
How does the covalent attachment of an m7G cap to circular mRNA (cap-circ mRNA) affect its ability to produce proteins compared to IRES-containing circular mRNAs?
What is the benefit of incorporating N1-methylpseudouridine (m1Ψ) modification into cap-circ mRNA, and how does it impact translation and immunogenicity in mice?
Explain the mechanism by which the non-covalent attachment of an m7G cap-containing oligonucleotide (cORN) enhances translation from circular mRNA.
What evidence suggests that the translation initiation process of cap-circ mRNA utilizes conventional factors for cap-dependent translation? Provide at least two examples.
How did in vivo studies in mice demonstrate the potential advantages of cap-circ m1Ψ mRNA compared to linear m1Ψ mRNA and IRES-containing circular mRNA in terms of protein expression and immune response?
Describe the concept of rolling circle-type translation of circular mRNA, and how can cORNs facilitate this process?
Explain how the cORN-mediated system can be utilized for endogenous RNA detection, providing an example from the study involving lncRNAs or mRNAs.
What are the key advantages of the internal cap-initiated translation (ICIT) strategies presented in this study compared to IRES-based methods for circular mRNA translation?
Quiz Answer Key
The primary challenge for therapeutic application of circular mRNA is the lack of a 5' cap structure, which is essential for efficient recruitment of the translation machinery. Traditional methods utilize Internal Ribosome Entry Sites (IRES) to initiate translation in a cap-independent manner by directly recruiting ribosomes.
The first design involves covalently attaching an N7-methylguanosine (m7G) cap to circular mRNA through a branching structure (cap-circ mRNA). The second design uses non-covalent attachment of an m7G cap by hybridizing the circular mRNA with an m7G cap-containing oligonucleotide (cORN).
Covalently capped circular mRNA (cap-circ mRNA) exhibits significantly more efficient protein production than IRES-containing circular mRNAs. This is because the m7G cap directly recruits the canonical translation machinery, leading to higher levels of protein expression.
Incorporating N1-methylpseudouridine (m1Ψ) modification into cap-circ mRNA reduces its acute immunostimulatory effect in mice while maintaining high translation ability. This modification helps the mRNA evade recognition by the innate immune system.
The non-covalent attachment of a cORN to circular mRNA through hybridization places an m7G cap structure in proximity to the start codon. This allows the circular mRNA to recruit the cap-dependent translation machinery, thereby enhancing protein expression.
Evidence for cap-dependent translation of cap-circ mRNA includes its sensitivity to hippuristanol, an eIF4A inhibitor, and the significant reduction in translation upon siRNA-mediated knockdown of eIF4E, the cap-binding protein. Additionally, the translation was blocked by oligoribonucleotide A30, a PABP competitor.
In vivo studies showed that subcutaneously administered cap-circ m1Ψ mRNA resulted in higher and more persistent protein expression compared to linear m1Ψ mRNA. In both subcutaneous and intravenous routes, cap-circ m1Ψ mRNA outperformed HRV-B3-IRES-containing circ mRNA, and also demonstrated a suppressed innate immune response compared to unmodified IRES-circ mRNA.
Rolling circle-type translation in circular mRNA occurs when translation initiation happens on a circular RNA without a stop codon, leading to continuous translation and the production of long proteins with tandem repeats. Capped cORNs can facilitate this by enhancing the rate-limiting initiation step on the circular mRNA.
The cORN-mediated system can detect endogenous RNA by designing an uncapped circular reporter mRNA with a sequence complementary to the target RNA. Upon hybridization with the capped target RNA (e.g., a lncRNA like HULC or mRNA like hACTB), the circular mRNA can "borrow" the cap structure and initiate translation of a reporter protein, indicating the presence of the target RNA.
• 10. ICIT strategies offer several advantages over IRES-based methods, including higher translation efficiency (in the case of covalent capping), compatibility with base modifications like m1Ψ without compromising translation, smaller molecular size of the circular mRNA, and the applicability to developing translational switches for RNA detection and cell-type specific translation.
3. Essay Questions
Essay Format Questions
Discuss the potential of internal cap-initiated translation (ICIT) as a next-generation technology for mRNA therapeutics and vaccines, highlighting its advantages and potential challenges compared to existing approaches.
Critically evaluate the two distinct ICIT strategies presented in the study—covalent and non-covalent cap attachment—considering their mechanisms, efficiencies, complexities, and suitability for different applications.
Analyze the role of mRNA stability and immunogenicity in the development of circular RNA therapeutics, and explain how the molecular designs presented in the study address these critical factors.
Explore the broader implications of the cORN-mediated system beyond enhancing circular mRNA translation, focusing on its utility in RNA diagnostics, translational control, and potential for developing novel therapeutic strategies based on endogenous RNA interactions.
Based on the findings of this study, propose future research directions that could further optimize internal cap-initiated translation of circular mRNA and expand its applications in biomedicine.
4. Glossary of Key Terms
Glossary of Key Terms
Circular mRNA (circRNA): A type of RNA molecule in which the 3' end is covalently joined to the 5' end, forming a continuous loop. This structure provides increased stability compared to linear mRNA.
Internal Ribosome Entry Site (IRES): An RNA sequence within an mRNA molecule that can directly recruit ribosomes and initiate translation in a cap-independent manner.
N7-methylguanosine (m7G) cap: A modified guanine nucleotide added to the 5' end of most eukaryotic mRNAs. It plays a crucial role in ribosome recruitment and translation initiation.
Internal Cap-Initiated Translation (ICIT): A mechanism of translation initiation on circular mRNA that utilizes an m7G cap positioned internally, either covalently or non-covalently.
cap-circ mRNA: Circular mRNA with an m7G cap covalently attached through a branching structure.
N1-methylpseudouridine (m1Ψ): A modified uridine nucleotide commonly incorporated into therapeutic mRNAs to reduce immunogenicity and enhance translation.
m7G cap-containing oligonucleotide (cORN): A short RNA molecule containing an m7G cap that is designed to hybridize to a target circular mRNA, non-covalently providing a cap for translation initiation.
Rolling circle translation: A process of continuous translation that can occur on circular mRNA lacking a stop codon, resulting in the production of long polypeptide chains with repeating units.
Long non-coding RNA (lncRNA): RNA molecules longer than 200 nucleotides that do not code for proteins but can have various regulatory functions within the cell.
HeLa cells: A human cervical cancer cell line widely used in biological research.
Lipid nanoparticles (LNPs): Small spherical vesicles made of lipids used to encapsulate and deliver nucleic acids (like mRNA) into cells.
Enzyme-linked immunosorbent assay (ELISA): A plate-based assay technique for detecting and quantifying substances like peptides, proteins, antibodies, and hormones.
Eukaryotic initiation factor 4A (eIF4A): An RNA helicase involved in unwinding secondary structures in the 5' UTR of mRNA to facilitate ribosome scanning.
eIF4E: The cap-binding protein that initiates the recruitment of the ribosome to the 5' cap of mRNA.
Poly A-binding protein (PABP): A protein that binds to the poly(A) tail at the 3' end of mRNA and interacts with eIF4F to circularize the mRNA and enhance translation.
siRNA: Small interfering RNA, a type of double-stranded RNA that can induce the degradation of complementary mRNA molecules, leading to gene silencing.
UTR: Untranslated region, the regions at the 5' and 3' ends of an mRNA molecule that are not translated into protein but contain regulatory elements.
CDS: Coding sequence, the region of an mRNA molecule that contains the genetic information for protein synthesis.
IVT: In vitro transcription, the process of synthesizing RNA molecules from a DNA template using RNA polymerase in a cell-free system.
RNase R: A 3' to 5' exonuclease that degrades linear RNA molecules but not circular RNA molecules, often used to confirm RNA circularity.
5. Timeline of Main Events
Pre-November 5th, 2024:
Development of IRES-containing circular mRNAs: Prior to this study, circular RNAs relied on Internal Ribosome Entry Sites (IRES) to initiate translation in a cap-independent manner. However, this method had limitations in efficiency.
Previous work on chemical capping: A previously reported method was developed for efficiently introducing the m7G cap structure into the monophosphate group of synthetic oligonucleotides using m7GDP imidazolide.
Development of a hydrophobic, photocleavable tag-containing chemical phosphorylation reagent: This was developed for purifying monophosphorylated RNA.
Discovery of endogenous circular RNAs and their translation: In the past decade, a significant number of endogenous circular RNAs have been discovered, and some are known to be translated.
Development of SINEUP lncRNAs: These long non-coding antisense RNAs were discovered to control the translation of target mRNAs through an embedded SINEB2 repeat.
Development of "translation activating RNAs": These RNAs, comprising antisense sequences to the 3' UTR of target mRNAs and IRES sequences, were developed to promote translation.
Early Stage of the Study:
Design of cap-circ mRNA: The researchers designed a circular mRNA with a covalently attached N7-methylguanosine (m7G) cap through a branching structure.
Synthesis of capped RNA oligos: Chemically synthesized oligoribonucleotides containing the cap on a branched strand were created using m7GDP imidazolide.
Synthesis of IVT-RNA: In vitro transcribed (IVT) RNA containing the coding sequence for NanoLuc luciferase (Nluc) was produced.
Ligation of RNA fragments: The capped RNA oligos and IVT-RNA were ligated using T4 RNA ligase 2 guided by splint DNAs to create cap-circ mRNA.
Confirmation of circular mRNA formation: Gel electrophoresis, reverse transcription, Sanger sequencing, and RNase R digestion were used to confirm the successful ligation and circularity of the synthesized RNAs.
Initial testing of cap-circ mRNA translation: The translation activity of cap-circ mRNA was measured in cultured HeLa cells, showing a significant increase in protein expression compared to uncapped circular RNAs.
Optimization of branch strand: The position and length of the capped branched strand were investigated to determine their impact on translation efficiency. A 6-nucleotide branch provided the highest activity.
Comparison with linear mRNA: Cap-circ mRNA showed more durable protein expression over time in cells and in vivo compared to linear mRNA, likely due to its increased stability.
Testing with m1Ψ modification: Incorporation of N1-methylpseudouridine (m1Ψ) into cap-circ mRNA further increased protein synthesis efficiency and reduced immunostimulatory effects in mice.
Comparison with IRES-containing circular mRNAs: Cap-circ mRNA showed comparable or superior translation activity to circular mRNAs containing EMCV or HRV-B3 IRES, and unlike IRES-driven translation, it was compatible with m1Ψ modification.
Testing with eGFP reporter: The cap-circ mRNA design was successfully applied to express enhanced green fluorescent protein (eGFP).
Stability studies: m1Ψ-modified cap-circ RNA showed greater in-cell stability compared to linear m1Ψ RNA and non-modified IRES-circ RNA.
Mechanism of translation initiation: Experiments using hippuristanol (eIF4A inhibitor), eIF4E knockdown, and oligoribonucleotide A30 (PABP competitor) indicated that cap-circ mRNA translation relies on conventional cap-dependent translation factors (eIF4E, eIF4A, PABP).
In Vivo Studies:
LNP encapsulation and administration: Cap-circ m1Ψ mRNA, IRES-circ mRNA, and linear m1Ψ mRNA were encapsulated in lipid nanoparticles (LNPs) and administered intravenously (IV) and subcutaneously (SC) to mice.
Nluc expression monitoring: In vivo imaging (IVIS) was used to monitor the expression of Nluc encoded by the delivered mRNAs. Cap-circ m1Ψ mRNA showed higher or comparable expression to linear mRNA and outperformed IRES-circ mRNA. Subcutaneous administration showed more sustained expression from cap-circ mRNA.
Acute immune response assessment: ELISA assays showed that m1Ψ-modified cap-circ and linear mRNAs suppressed the innate immune response compared to unmodified IRES-circ mRNA.
Testing with short peptide-encoding mRNA: A smaller cap-circ mRNA encoding a HiBiT peptide showed prolonged and increased expression in the liver and spleen compared to its linear counterpart at later time points.
Development of Non-Covalent ICIT (cORNs):
Design and synthesis of capped ORNs (cORNs): m7G cap-modified oligoribonucleotides complementary to circular mRNA were designed and synthesized.
Testing cORN-mediated translation: Pre-annealing cORNs to circular mRNA enhanced translation even without IRES or capped branch strands, with the most distant hybridization site from the start codon showing the highest enhancement.
Optimization of cORNs: The length of the complementary sequence and chemical modifications (non-nucleotide linkers, 3' cap, poly-A tail) were explored to further enhance translation activation. Poly-A-tailed cORNs showed improved efficacy, especially for circular mRNAs lacking a poly-A tail.
Comparison of covalent and non-covalent ICIT: Covalent ICIT (cap-circ mRNA) was found to be more efficient than non-covalent ICIT (cORNs) in driving protein expression.
Combination with IRES: cORNs did not enhance the translation of HRV-B3-IRES-containing circular mRNA.
Advanced Applications of cORNs:
Rolling circle translation: cORNs were shown to facilitate rolling circle-type translation of circular mRNA with continuous reading frames, increasing the production of tandemly linked proteins.
Endogenous RNA detection: The cORN concept was applied to detect endogenous RNAs (lncRNAs and mRNAs) by designing circular reporter mRNAs that hybridize to the target capped RNA and initiate translation using the target RNA's cap.
Detection of HULC lncRNA: A circular mRNA complementary to HULC lncRNA showed increased translation in the presence of exogenous and endogenously expressed HULC RNA.
Detection of hACTB mRNA: Circular mRNAs with complementary regions to hACTB mRNA showed reduced translation upon hACTB knockdown, indicating that endogenous hACTB mRNA promotes their translation. Chemical modifications in the hybridization region further enhanced this effect.
November 5th, 2024:
Date the article was posted.
Cast of Characters (Principle People Mentioned):
Hiroshi Abe: Researcher at Nagoya University and co-founder of Crafton Biotechnology. A lead author and inventor on the patent application. Conceptualized and designed the study, and wrote the manuscript.
Kosuke Fukuchi: Researcher at Nagoya University. Performed chemical synthesis and biochemical experiments.
Yuko Nakashima: Researcher at Nagoya University. Performed chemical synthesis and biochemical experiments. An inventor on the patent application.
Naoko Abe: Researcher at Nagoya University. A lead author and inventor on the patent application. Conceptualized and designed the study, performed chemical synthesis and biochemical experiments, and wrote the manuscript.
Seigo Kimura: Researcher at Nagoya University. Designed and performed biochemical and animal experiments, and wrote the manuscript.
Fumitaka Hashiya: Researcher at Nagoya University. Performed biochemical experiments and is an inventor on the patent application.
Yuichi Shichino: Researcher at RIKEN. Performed biochemical experiments and wrote the manuscript.
Satomi Sugiyama: Researcher at Nagoya University and co-founder of Crafton Biotechnology. Performed chemical synthesis and biochemical experiments. An inventor on the patent application.
Daisuke Kawaguchi: Researcher at Nagoya University. Performed chemical synthesis and biochemical experiments.
Masahito Inagaki: Researcher at Nagoya University. Performed chemical synthesis and is an inventor on the patent application.
Zheyu Meng: Researcher at Nagoya University. Performed chemical synthesis and biochemical experiments.
Shiryu Kajihara: Researcher at Nagoya University. Performed biochemical experiments.
Mizuki Tada: Researcher at Nagoya University. Performed chemical synthesis.
Satoshi Uchida: Researcher at Tokyo Medical and Dental University. Designed and performed biochemical and animal experiments, and wrote the manuscript.
Ting-Ting Li: Researcher at Nagoya University. Performed chemical synthesis.
Ramkrishna Maity: Researcher at Nagoya University. Performed chemical synthesis.
Yasuaki Kimura: Researcher at Nagoya University. Performed chemical synthesis and is an inventor on the patent application.
Shintaro Iwasaki: Researcher at RIKEN. Performed biochemical experiments and wrote the manuscript. Member of the Scientific Reports editorial board.
Ryoko Ogisu: Researcher at Nagoya University. Performed chemical synthesis and biochemical experiments.
Tairin Kawasaki: Researcher at Nagoya University. Performed chemical synthesis.
Junichi Tanaka: Provided hippuristanol (eIF4A inhibitor).
Nicholas T. Ingolia: Provided psiCHECK2-HCV IRES.
Tomoe Nishikawa, Tomoyo Tada, Susumu Tsutsumi, Yukari Kondo: Provided technical assistance with IVT and animal experiments (Nagoya University).
Karikó, K. & Weissman, D.: Authors of studies on the suppression of RNA recognition by Toll-like receptors through nucleoside modification (pseudouridine).
Pelletier, J. & Sonenberg, N.: Authors of studies on eukaryotic cap structure and ribosome recruitment.
Jaffrey, S.R.: Author of studies on highly efficient cellular expression of circular mRNA and mRNA aging.
Chen, R. et al.: Authors of a study on engineering circular RNA for enhanced protein production.
Qu, L. et al.: Authors of a study on circular RNA vaccines against SARS-CoV-2.
Wesselhoeft, R.A. et al.: Authors of a study on the reduced immunogenicity and extended translation of circular RNA in vivo.
Carrieri, C. et al.: Authors of a study on SINEUP non-coding RNA controlling Uchl1 translation.
Cao, Y. et al.: Authors of a study on RNA-based translation activators.
6. FAQ
Frequently Asked Questions about Internal Cap-Initiated Translation (ICIT) of Circular mRNA
1. What are the main limitations of using circular mRNA for therapeutic applications, and how does this study address them?
Circular mRNA has emerged as a promising RNA therapeutic strategy due to its enhanced stability and reduced immunogenicity compared to linear mRNA. However, a key challenge has been its lower translation efficiency, as circular RNAs lack the 5' cap structure that is crucial for efficient ribosome recruitment in eukaryotic cells. This study introduces two novel molecular designs to overcome this limitation by enabling internal cap-initiated translation (ICIT). The first design involves covalently attaching an m7G cap to a circular mRNA through a branching structure (cap-circ mRNA), while the second design uses a non-covalent approach where an m7G cap is delivered by a complementary oligonucleotide that hybridizes to the circular mRNA (cORN-mediated ICIT). These advancements aim to boost protein production from circular mRNA, making it more suitable for therapeutic and diagnostic applications.
2. How does the covalently linked cap-circ mRNA enhance protein translation compared to traditional methods for circular mRNA translation?
Traditionally, circular mRNA translation relies on internal ribosome entry sites (IRESs), which recruit ribosomes in a cap-independent manner. However, IRES-mediated translation is generally less efficient than cap-dependent translation. The cap-circ mRNA design introduces an m7G cap structure directly onto the circular RNA, allowing it to be recognized by the eukaryotic translation initiation factor eIF4F, similar to linear mRNA. This study demonstrates that cap-circ mRNA can achieve protein expression levels two to three orders of magnitude higher than uncapped circular mRNA and often surpasses the efficiency of IRES-containing circular mRNAs. Furthermore, the translation from cap-circ mRNA was shown to be dependent on key cap-dependent translation factors like eIF4E and eIF4A, as well as PABP, indicating a mechanism similar to canonical translation initiation.
3. What role does N1-methylpseudouridine (m1Ψ) modification play in the context of cap-circ mRNA?
N1-methylpseudouridine (m1Ψ) modification is a common strategy used in mRNA therapeutics to reduce innate immune responses. This study found that incorporating m1Ψ into cap-circ mRNA not only lowered the acute immunostimulatory effect in mice but also surprisingly enhanced protein synthesis efficiency by nearly sixfold compared to unmodified cap-circ mRNA. This combination is particularly beneficial for vaccine and therapeutic applications where minimizing inflammation while maintaining high protein production is crucial. Interestingly, the study also noted that m1Ψ modification significantly attenuated the translation driven by IRES-containing circular mRNA, highlighting a potential advantage of the cap-circ mRNA approach when combined with this modification.
4. How does the non-covalent, cORN-mediated ICIT strategy work to enhance circular mRNA translation?
The second approach involves designing short, m7G cap-modified oligoribonucleotides (cORNs) that are complementary to a region within the circular mRNA, ideally near the start codon. When these cORNs hybridize to the circular mRNA, they effectively present an m7G cap in close proximity to the ribosome binding site, thereby facilitating recruitment of the translation machinery. This study showed that cORN hybridization could enhance translation from circular mRNAs by one to two orders of magnitude, even in the absence of IRES elements or capped branch strands. The translation enhancement was dependent on the presence of the cap on the cORN and its ability to hybridize to the target circular mRNA.
5. What are the potential advantages of the non-covalent cORN-mediated ICIT approach for therapeutic and diagnostic applications?
The cORN-mediated ICIT strategy offers several potential advantages. Firstly, it simplifies the design and synthesis of the circular mRNA itself, as it does not require the incorporation of a branched structure or an IRES element for efficient translation. The translational control is instead provided by the separate, chemically synthesized cORN. This modularity allows for the potential design of translation switches that can be activated in the presence of specific capped RNAs, such as disease-specific long non-coding RNAs (lncRNAs). The study demonstrated the feasibility of this concept by showing that a circular reporter mRNA could be designed to hybridize to capped lncRNAs (like HULC) or endogenous mRNAs (like β-actin), leading to protein production in a hybridization-dependent manner. This opens avenues for novel diagnostic tools and cell-type selective mRNA therapeutics.
6. How does the size of the circular mRNA in the ICIT strategies compare to IRES-containing circular mRNAs, and why is this significant?
The ICIT strategies, particularly the covalently capped cap-circ mRNA, allow for the creation of smaller circular mRNAs compared to those relying on IRES elements. IRES sequences are typically several hundred bases long, whereas the capped branch used in this study was significantly shorter (e.g., a 54-nt fragment resulting in a 604-nt total length for an Nluc-encoding circular mRNA). This smaller size can be advantageous for several reasons: it may improve packaging and delivery efficiency, potentially reduce off-target effects, and might influence the overall stability and translational kinetics of the mRNA. The study even demonstrated the successful design of a 200-nt cap-circ mRNA encoding a short peptide, highlighting the potential for minimal mRNA constructs.
7. What are the in vivo findings regarding the efficacy and immunogenicity of the ICIT-based circular mRNAs compared to linear and IRES-containing circular mRNAs?
In vivo studies in mice demonstrated that m1Ψ-modified cap-circ mRNA exhibited comparable or superior protein expression to optimized IRES-containing circular mRNA, depending on the route of administration (intravenous vs. subcutaneous). Notably, after subcutaneous administration, the cap-circ mRNA showed higher and more sustained protein expression than linear mRNA, potentially due to its increased stability. Furthermore, the m1Ψ-modified cap-circ mRNA showed suppressed innate immune responses, similar to m1Ψ-modified linear mRNA, and significantly lower than unmodified IRES-containing circular mRNA. These findings support the potential of ICIT-based circular mRNAs for safe and effective in vivo applications.
8. What are the future directions and potential implications of this research for the field of RNA therapeutics and diagnostics?
This research introduces a versatile framework for enhancing protein production from circular mRNA through internal cap-initiated translation, addressing a major bottleneck in the field. The two distinct strategies, covalent cap attachment and non-covalent cap delivery via cORNs, offer complementary advantages for different applications. The ability to combine cap-dependent translation with nucleoside modifications and to achieve efficient translation with smaller RNA constructs has significant implications for the development of more effective and safer mRNA vaccines and therapeutics. The cORN-mediated system also opens exciting new possibilities for creating RNA-based sensors and translation switches that can respond to specific cellular RNAs, paving the way for novel diagnostic tools and targeted therapies. Future work may focus on further optimizing these ICIT designs, elucidating the precise mechanisms of cORN-mediated translation, and exploring their applications in a wider range of biological contexts.
7. Table of Contents with Timestamps
Contents: Circular mRNA and Internal Cap-Initiated Translation
00:00 - Introduction to Circular mRNA A welcome to the Deep Dive podcast and introduction to the topic of circular mRNA and its potential in RNA therapeutics.
00:18 - The Paper's Title and Significance Discussion of the recently published paper "Internal Cap-Initiated Translation Provides Efficient Protein Production from Circular mRNA" and its implications.
00:32 - Stability Advantage of Circular mRNA Explanation of why circular mRNA is more stable than traditional linear mRNA and how this leads to potentially more potent treatments.
00:56 - The Challenge: The Missing Cap Description of the key problem with circular mRNA - the absence of the M7G cap that allows mRNA to enter the cell's protein-making machinery.
01:28 - ICIT: The Solution Introduction to Internal Cap-Initiated Translation (ICIT) as the innovation developed by Nagoya University researchers.
01:46 - Two Approaches to ICIT Detailed explanation of the two methods developed: Covalent ICIT and Non-covalent ICIT.
02:20 - Results: Dramatic Increase in Protein Production Discussion of the impressive experimental results showing 50-100 fold increases in protein production.
03:05 - In Vivo Testing Description of how the covalent ICIT method performed in mice, demonstrating higher and more consistent protein production.
03:33 - M1 Modification for Immune System Evasion Explanation of how combining cap circ mRNA with an M1 modification reduces immune system reaction.
04:00 - Deep Dive into CORN Technology Detailed exploration of the non-covalent approach using CORN (Capped Oligoribonucleotides).
04:37 - Surprising Findings About CORN Positioning Discussion of counterintuitive discoveries regarding optimal placement of the CORN on circular mRNA.
05:24 - Chemical Modifications to Enhance Performance Examination of various chemical modifications tested to improve CORN stability and binding.
06:13 - Rolling Circle Translation Explanation of the groundbreaking discovery of rolling circle translation, where multiple protein copies can be produced from a single mRNA.
06:56 - RNA Detection Applications Discussion of how CORN can be adapted to sense and detect specific RNAs within cells.
07:51 - Cancer Diagnostic Potential Exploration of how this technology could be used to detect cancer-associated RNAs like HULC.
08:14 - Detecting Endogenous RNAs Description of how the system can detect naturally produced RNAs within cells.
08:54 - Challenges and Limitations Assessment of the hurdles facing this technology before clinical applications can be realized.
10:19 - Broader Implications for RNA Therapeutics Discussion of the versatility and potential applications beyond disease treatment.
10:53 - Future of Personalized Medicine Consideration of how these technologies could enable more tailored treatments based on individual RNA profiles.
11:24 - Conclusion Summary of the significance of this research and its potential to reshape the future of medicine.
8. Index with Timestamps
Actin (beta-), 08:32, 08:37
Adenine nucleotides, 05:41
Artifact, 06:44
BNA bases, 05:55
Beta-actin mRNA, 08:32, 08:37
Binding affinity, 05:34
Biomaterials, 07:17
Biomanufacturing, 11:09
Cancer, 07:59, 08:03
Cap, 01:01, 01:02, 01:09, 01:15, 02:00, 02:20, 02:33, 02:41, 07:41
Capped circular mRNA, 03:43
Chemical modifications, 04:24, 05:24, 05:25, 05:41, 05:55
Circular mRNA, 00:13, 00:32, 00:36, 00:41, 01:10, 01:15, 02:00, 02:17, 02:20, 03:43, 07:41, 11:17, 11:18, 11:47
Covalent ICIT, 01:52, 01:53, 03:12, 09:14
Degradation, 05:59, 10:08
Delivery (challenges), 10:04
Diagnostics, 08:03, 10:36, 10:42
Diseases, 03:33, 07:06, 10:29, 10:32, 10:42, 11:47, 11:58
FLAG-EGF, 06:44
HULC, 07:57, 08:00
ICIT (Internal Cap-Initiated Translation), 01:40, 01:41, 01:52, 09:14
Immune system, 03:43, 03:48
Linear mRNA, 00:37, 00:40, 01:01, 02:04, 03:22
LNA bases, 05:55
M1, 03:43, 03:43
M7G, 01:01, 01:02, 01:10, 02:33, 02:41
Mice, 03:12
Nagoya University, 01:28, 11:28, 11:33
Non-covalent ICIT, 01:52, 01:53, 02:25, 09:34, 09:43
Nucleotides, 05:41, 05:55
Off-target effects, 09:53
Poly-A tail, 05:41
Position (of CORN), 04:42, 04:48
Protein, 02:17, 02:20, 02:54, 02:58, 03:18, 03:22, 04:44, 04:49, 05:07, 05:36, 06:29, 06:44, 06:49, 06:51, 07:08, 09:53, 11:07, 11:09
RNA detection, 07:51, 08:43
Rolling circle translation, 06:13, 06:15, 06:18, 06:23, 06:44, 06:49, 11:07
Scaling up, 09:22
Sense RNAs, 07:32, 07:35
Stability, 05:34, 09:48
Theranostics, footnote
Translation, 01:40, 01:41, 02:20, 02:41, 02:44, 04:49, 05:07, 06:13, 06:15, 06:18, 06:23, 06:29, 06:44, 06:49, 06:51, 07:41, 09:48, 11:07
VIP pass, 01:09
9. Poll
10. Post-Episode Fact Check
The content of this podcast episode appears to accurately represent cutting-edge research on circular mRNA technology. Key claims that can be verified include:
1. ✓ The description of circular mRNA being more stable than linear mRNA is accurate. Circular RNA molecules lack free ends that cellular exonucleases typically attack, leading to longer persistence in cells.
2. ✓ The explanation of the M7G cap being necessary for efficient translation is correct. In standard mRNAs, this 5' cap is essential for ribosome recruitment.
3. ✓ The general concept of Internal Cap-Initiated Translation (ICIT) and the two approaches (covalent and non-covalent) align with emerging research in RNA therapeutics.
4. ✓ The reported fold-increases in protein production (50-100x) are within the range of improvements reported in recent scientific literature for circular RNA optimization.
5. ✓ The potential applications in diagnostics, including detection of cancer-associated RNAs like HULC, are consistent with the current direction of RNA technology research.
The challenges mentioned (manufacturing complexity, delivery mechanisms, scaling) accurately reflect the current hurdles facing RNA therapeutic development. Overall, the podcast presents a scientifically sound explanation of circular mRNA technology and its potential.
11. Image (3000 x 3000 pixels)
12. Mind Map